The Eckart Young Mirsky Theorem States the following. Suppose $A \in \mathbb{R}^{m \times n}$
$$ A = U \Sigma V^{T} \tag{1} $$
then if we take a rank k approximation of the matrix using the SVD
$$A_{k} = \sum_{i=1}^{k} \sigma_{i}u_{i}v_{i}^{t} \tag{2} $$
the difference between them is given as
$$ \| A - A_{k} \|_{2} = \bigg\| \sum_{i=k+1}^{n} \sigma_{i}u_{i} v_{i}^{t} \bigg\| = \sigma_{k+1} \tag{3}$$
The best rank k approximation is when the matrix has the given rank k. This is from this expression. If $A= A_{k}$ our minimization expression will be minimized.
I am not sure how $D$ is going to help you better approximate the matrix $\Sigma $. I am sorry about the confusion with notation.
Right, if $k_{2}$ is less than $k$ it is actually not good
$$ \| A + D\| \leq \| A \| + \|D\| \tag{4}$$
and we can approximate both of these. The norm is
$$ \| A\|_{2} = \sqrt{\lambda_{max}(A^{*}A)} = \sigma_{max}(A) = \sigma_{1} \tag{5}$$
the 2-norm is the maximum singular value.
$$ \| A + D\| \leq \sigma_{max}(A) + \sigma_{max}(D) \tag{6}$$
So from above, you have
$$ \| A -\Sigma_{1} \|_{2} = \sigma_{k_{1}+1} \tag{7} $$
$$ \| A- \Sigma_{2} \|_{2} - \|D \|_{2} \leq \| A - \Sigma_{2} -D \|_{2}\tag{8} $$
Then
$$ \| A- \Sigma_{2}\|_{2} = \sigma_{k_{2}+1} \\ \|D\|_{2} = \sigma_{max}(D) \tag{9}$$
$$ \sigma_{k_{1}+1} - \sigma_{k_{2}+1} - \sigma_{max}(D) \leq \|A-\Sigma_{1} \|_{2} -\|A-\Sigma_{2} -D\|_{2} \tag{10}$$
Note that singular values are ordered so $\sigma_{k_{1}+1} \geq \sigma_{k_{2}+1}$ if $k_{1} \geq k_{2}$
Numerical Simulation
I think you wanted a numerical simulation to support your argument so I am going to create the Python code to show you why. I already answered a similar question about SVDs earlier. So I will use that code to show you.
Say we have our covariance matrix or really any matrix $A$ and it has a rank $\alpha$,
import numpy as np
import matplotlib.pyplot as plt
import math
def gen_rank_k(m,n,k):
# Generates a rank k matrix
# Input m: dimension of matrix
# Input n: dimension of matrix
# Input k: rank of matrix
vec1 = np.random.rand(m,k)
vec2 = np.random.rand(k,n)
rank_k_matrix = np.dot(vec1,vec2)
return rank_k_matrix
m=10
n=m
alpha = 7
A = gen_rank_k(m,n,k)
Now we have $k_{2} \leq k_{1} \leq \alpha $
u, s, vh = np.linalg.svd(A, full_matrices = False)
x = np.linspace(1,10,10)
plt.plot(x,s)
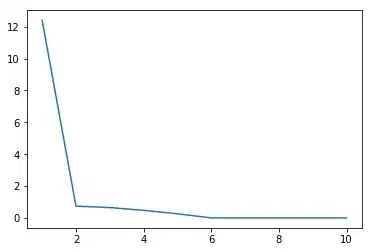
This is a plot of the singular values of $A$
now for the two low rank approximations of $A$ choose $k_{1} = 5, k_{2} = 3$
def low_rank_k(u,s,vh,num):
# rank k approx
u = u[:,:num]
vh = vh[:num,:]
s = s[:num]
s = np.diag(s)
my_low_rank = np.dot(np.dot(u,s),vh)
return my_low_rank
my_rank_k1 = low_rank_k(u,s,vh,k1)
my_rank_k2 = low_rank_k(u,s,vh,k2)
my_diagonal_matrix = np.diag(np.random.rand(10))
error1 = np.linalg.norm(A - my_rank_k1)
error2 = np.linalg.norm(A- my_rank_k2 - my_diagonal_matrix)
