Let's say I have dataset within the following pandas dataframe format with a non-standard timestamp column without datetime format as follows:
+--------+-----+
|TS_24hrs|count|
+--------+-----+
|0 |157 |
|1 |334 |
|2 |176 |
|3 |86 |
|4 |89 |
... ...
|270 |192 |
|271 |196 |
|270 |251 |
|273 |138 |
+--------+-----+
274 rows × 2 columns
The dataset shape is $274*2$ and contains the first column of timestamp and 2nd column of numerical statistical count as the label, and I want to train some ML\DL regression models and do out-of-sample prediction (predicting beyond the training dataset) over future data (unseen test-set). I have already implemented RF regression within sklearn pipeline() after splitting data with the following strategy for 274 records:
- split data into [training-set + validation-set] Ref. e.g. The first 200 records [160 +40]
- keeping unseen [test-set] hold-on for final forecasting e.g. The last 74 records (after 200th rows\event)
#print(train.shape) #(160, 2)
#print(validation.shape) #(40, 2)
#print(test.shape) #(74, 2)

#ِDataset matrix is formed
def create_dataset(df , lookback=1):
data = np.array(df.iloc[:, :-1])
label = np.array(df.iloc[:, -1])
#create X_train and Y_train and MinMaxScaler on Y_train
X = list()
Y = list()
for i in range(lookback , len(data)):
X.append(data[i-lookback:i])
Y.append(label[i])
X = np.array(X)
Y = np.array(Y)
Y=np.expand_dims(Y,-1)
return X,Y
Lookback period
lookback = 5
X_train, Y_train = create_dataset(train, lookback)
X_val, Y_val = create_dataset(validation, lookback)
X_test, Y_test = create_dataset(test, lookback)
print(X_train.shape , Y_train.shape) #(155, 5, 1) (155, 1)
print(X_val.shape, Y_val.shape) #(35, 5, 1) (35, 1)
print(X_test.shape, Y_test.shape) #(69, 5, 1) (69, 1)
- transform a time series dataset into a supervised learning dataset posted by Jason Brownlee:
# Finalize the model and make a prediction for monthly births with random forest
from numpy import asarray
from pandas import read_csv
from pandas import DataFrame
from pandas import concat
from sklearn.ensemble import RandomForestRegressor
transform a time series dataset into a supervised learning dataset
def series_to_supervised(data, n_in=1, n_out=1, dropnan=True):
n_vars = 1 if type(data) is list else data.shape[1]
df = DataFrame(data)
cols = list()
input sequence (t-n, ... t-1)
for i in range(n_in, 0, -1):
cols.append(df.shift(i))
forecast sequence (t, t+1, ... t+n)
for i in range(0, n_out):
cols.append(df.shift(-i))
put it all together
agg = concat(cols, axis=1)
drop rows with NaN values
if dropnan:
agg.dropna(inplace=True)
return agg.values
load the dataset
series = read_csv('daily-total-female-births.csv', header=0, index_col=0)
values = series.values
transform the time series data into supervised learning
train = series_to_supervised(values, n_in=6)
split into input and output columns
trainX, trainy = train[:, :-1], train[:, -1]
fit model
model = RandomForestRegressor(n_estimators=1000)
model.fit(trainX, trainy)
construct an input for a new prediction
row = values[-6:].flatten()
make a one-step prediction
yhat = model.predict(asarray([row]))
print('Input: %s, Predicted: %.3f' % (row, yhat[0]))
Qs:
Q1: What is the difference between these two approaches? (lookback period Vs transform a time series dataset into a supervised learning dataset)
I also tried in meantime walk_forward_validation() approach inspired from this post:
Walk forward validation is a method for estimating the skill of the model on out of sample data. We contrive out of sample and each time step one out of sample observation becomes in-sample. We can use the same model in ops, as long as the walk-forward is performed each time a new observation is received.
My observation shows no difference between using:
- just
series_to_supervised(STS) walk_forward_validation(WFV) &series_to_supervised(STS) comparing outputs, especially MAE results, but a bit in quality of forecasting up & downs(could be due to weights during training):
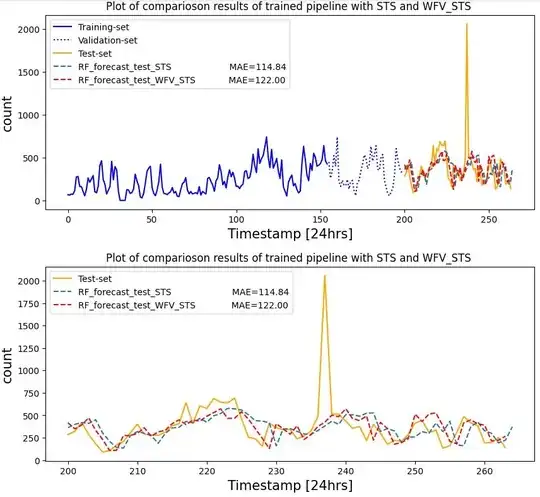
Q2: How possibly can I reflect\compare prediction results when data split size based on my strategy has undergone of above methods, e.g. Look back period while lookback = 5? (I already missed 5 records, and in practice leads delay in forecasting the right point)
# The first 200 records slice for training-set and validation-set
df200=df[:200]
The rest records = 74 events (after 200th event) kept as hold-on unseen-set for forcasting
test = df[200:] #test
Split the data into training and testing sets
from sklearn.model_selection import train_test_split
train, validation = train_test_split(df200 , test_size=0.2, shuffle=False) #train + validation
#print(train.shape) #(160, 2)
#print(validation.shape) #(40, 2)
#print(test.shape) #(74, 2)
```